1-Two levels of curation
There are two levels of curation:
1-Curation of the ontology which includes curation of the term name, synonyms, definition, cross-references and relationships. Based on the curation input using RACE-PRO or a tab-delimited file, a file with preliminary stanzas in OBO format is generated by an in-house script (OBO Maker). The file is checked via the OBO release manager (OORT) is done before posting.
2-Annotation of a PRO term based on the experimental knowledge supported by literature.
The annotation of the PRO terms is distributed in a tab delimited file called PAF (https://proconsortium.org/download/documents/PAF_guidelines.pdf). This file follows closely the format of the gene ontology association file (GAF). Annotation is added using RACE-PRO interface o via a template PAF file.
2-Ontology structure

PRO Hierarchy: Illustration of PRO categories and relation to external resources. Categories (aka metaclasses) are listed along the top, with example terms for IRF5 (interferon regulatory factor 5) shown directly below. Circles denote protein terms, while squares denote protein complex terms. For each, blue indicates terms that are defined in an organism-agnostic way, green indicates terms that are organism-specific. Solid lines show is_a relationships and broken lines show has_component relationships (note that, though the complexes in this example have components in the modification category, this need not be the case). Filled circles and filled squares indicate how UniProtKB and GO, respectively, fit into the hierarchy. Not all terms and relationships are shown. IRF, interferon regulatory factor; Phos, phosphorylated; m, mouse; h, human; iso, isoform; BMv, bone marrow variant.
2.1 -PRO IDs
PRO reuses external ontology identifiers whenever feasible. Thus, if a required term is already defined in another ontology, that term is used by PRO without changing its identifier (that is, we do not create an identical term and assign a PRO identifier to it). However, much information in the protein field is available in the UniProtKB database, with established and stable accessions. Therefore, when we incorporate UniProtKB entries into the PRO 'organism-gene' category, in the interest of minimizing the minting of new identifiers, we use UniProtKB accession numbers directly, prefixing them with "PR:" as in the example PR:P19883 ("http://purl.obolibrary.org/obo/PR_P19883" for the URL). All such entries are cross-referenced back to the originating database, using "UniProtKB:" as the prefix, as in the example "UniProtKB:P19883" ("http://www.uniprot.org/uniprot/P19883" for the URL). In this way the database and ontological representations of P19883 are clearly distinguished. In the future, use of UniProtKB isoform identifiers (of the form P19883-1) will be adopted.
2.2-PRO Term Names
All PRO term names are lower case except for certain standard abbreviations such as DNA, ATP, GTP, etc. Term names are based on naming guidelines that follow nomenclature suggested by nomenclature committees, literature, and UniProtKB (http://www.uniprot.org/docs/nameprot), among others.
3-Curation of the ontology
3.1-ProEvo terms
ProEvo encompasses the categories: family and gene.
3.1.1 Category=family: Each PRO term at this level refers to protein products of a distinct gene family arising from a common ancestor. The leaf-most nodes at this level are usually families comprising paralogous sets of gene products (of a single or multiple organisms). For example, smad2 and smad3 both encode proteins that are TGF??receptor-regulated while smad1, smad5, and smad9 are all BMP receptor-regulated. Thus, "TGF-beta receptor-regulated smad protein" and "BMP receptor-regulated smad protein" are terms denoting distinct families. Note that this level collectively refers to any such grouping at any level of similarity. For example, the two families indicated above can merge into a "receptor-regulated smad protein" class and further
merge (with the protein products of smad4, smad6, and smad7) into the "smad protein" class (Fig1). No merging occurs beyond what can be done to account for the evolution of the entire, full length protein. That is, all proteins that can trace back to a common ancestor over the entire length of the protein are part of the same family.
3.1.2 Category=gene: Each PRO term at this level refers to the protein products of a distinct gene in a reference organism and the orthologs thereof. For example, "smad2" and "smad3" are two different genes, and therefore have two different PRO entries at the gene level of distinction. The protein products of what is recognized as smad2 in humans and what is recognized as smad2 in mouse thus fall under this single term. Thus, a single term at the gene-level distinction collects the protein products of a subset of orthologs for that gene. Gene-level distinction is the leaf-most node of the ProEvo part of PRO.
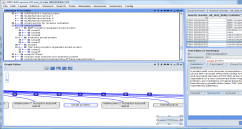
Fig1- Snapshot of the ProEvo part of the ontology for smad proteins
Curation of Category=family
Stanza:
[Term]
id: PR:<ID>
name: <name>
def: "A <parent> with core domain composition consisting of n copies of <Pfam domain name> (<Pfam domain ID>, and ….." [PRO:<curator initials>, PMID:<number>, <DBname>:<DB ID>]
comment: Category=family.
synonym: "<name>" <EXACT/RELATED/BROAD/NARROW> []
xref: <DB name>:<DB ID>
is_a: <parent id> ! <parent name>
Curation of these terms is part of the Protein Information Resource (PIR) core activities, however, the name and definitions of the terms can always be improved by collaboration with experts. Here is a brief description on how these terms are curated. The ProEvo classes are built based on the homologous and homeomorphic concepts [Wu et al, 2004], proteins sharing full-length sequence similarity and same domain architecture. These terms are automatically generated from the databases PIRSF [Wu et al, 2004] and PANTHER [Mi et al, 2006] based on the membership to the family and subfamily level for PIRSF and the subfamily level for PANTHER.
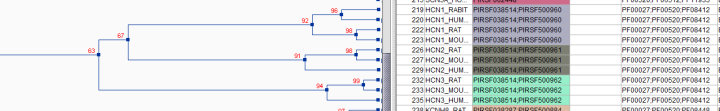
Fig .2- shows a partial result on the iterative BLAST Clust procedure on UniProtKB-Swiss-Prot proteins containing PF00520 (ion channel domain), showing the corresponding mapping to PIRSF and PANTHER, alongside the corresponding PRO terms generated. Note that the first cluster (1) shows a group for which there is no PIRSF subfamily level, but the PANTHER classification provides this level. In the second cluster (2), there is no current PANTHER classification for the group, then PIRSF subfamily level provides the granularity needed. The Pfam column gives information for the domain architecture of the proteins in the group, and 3) In the case a PIRSF subfamily and PANTHER subfamily map 1 to 1, only one term should be created with cross-ref to the two.
If the homeomorphic level coincides with the gene product level, the Category=gene should be assigned (like case 3 in Figure 2).
case 1

In the ontology:

case 2
In the ontology: 
case 3
In the ontology: 
Fig2- Iterative BLAST Clust procedure on UniProtKB-Swiss-Prot proteins containing PF00520 (ion channel domain), showing the corresponding mapping to PANTHER along with the ontology hierarchy.
PRO term names
PIRSF names may contain a "type" qualifier to distinguish among different families with the same name. Although this nomenclature is appropriate for PIRSF it is not for PRO ontology, so those names have to be resolved based on other knowledge. In the example shown above this particular family becomes the "reference" voltage-gated potassium channel alpha subunit, whereas the other type have some additional qualifier. One of these cases is the "voltage-gated potassium channel, alpha subunit, Eag/Elk/Erg types". The hallmark for this group is the presence of a PAS domain at the N-terminus, therefore, this protein class could be named instead "voltage-gated potassium channel, alpha subunit, with PAS domain".
This has to be looked at on an individual basis, with consultation with literature and experts.
Names are spelled out unless there is a widely spread acronym, like smad. All other names then are added as synonyms.
PRO term definitions
- Many ProEvo family definitions follow the formula:
"A <parent> that contains <number of copies> of <domain name 1> (<PFam ID 1>), <number of copies> of <domain name 2> (<PFam ID 2>), …. <number of copies> of <domain name n> (<PFam ID n>).
Example:
[Term]
id: PR:000002997
name: proto-oncogene SRC-like tyrosine-protein kinase
def: "A protein that contains one copy of the SH3 domain (PF00018), a copy of the SH2 domain (PF00017), and a copy of the Protein tyrosine kinase (PF07714) domain. Members of this class are cytoplasmic protein-tyrosine kinases believed to be involved in signal transduction." [PRO:CNA, PMID:7545532]
comment: Category=family.
synonym: "Src family tyrosine kinase" EXACT []
synonym: "SFK" RELATED []
xref: PIRSF:PIRSF000601
is_a: PR:000000001 ! protein
Add Pfam domain names as they are originally in the Sanger database that is first letter capitalized and with or without domain in the name. The Pfam ID will follow the name in parenthesis. If the word domain is not part of the Pfam domain then we will add it after the ID. If there are multiple copies of a domain, then add it in the form of ".. n copies of the ……(PFxxxx) domain".
- It is also permitted to refer to an outside authority, as in "A protein that is a translation product of some gene evolved from the last common ancestor of subfamily V of the transient receptor potential cation channels (TRPs)." The corresponding citation is required for this form.
- In the case of small families, the genes may be enumerated, as in "An LPAR1/S1PR1-like lysophospholipid receptor that is a translation product of the LPAR1, LPAR2, or LPAR3 gene."
- Subfamily definitions and gene level definitions should not repeat domain information.
- Subsequent sentences can give other useful information: functional characteristics, motifs, evolution, including gene expansions and contractions.
- For subfamily level, definitions usually are of the type:
"A parent term that add differentia"
example: PR:000000709 "A voltage-gated potassium channel alpha subunit that is a member of the subfamily A of voltage-gated potassium channels. The N-terminus is rich in basic residues and this region is critical for the N-type inactivation. Shaker-related voltage-gated potassium channels (Kv1 channels) play an important role in modulating electrical excitability of axons."
Cross-Ref
For ProEvo we provide the corresponding cross-reference to PIRSF and PANTHER as needed following the format:
xref: PANTHER:<PANTHER ID>
xref: PIRSF:<PIRSF ID>
Curation of Category=gene
Terms in the gene category comprise the largest metaclass within PRO.
Stanza:
[Term]
id: PR:<ID>
name: <name>
def: "A <parent> that is a translation product of the <organism> <gene symbol> gene or a 1:1 ortholog thereof." [PRO:<curator initials>, PMID:<number>, <DBname>:<DB ID>]
comment: Category=gene.
synonym: "<name>" <EXACT/RELATED/BROAD/NARROW> []
xref: <DB name>:<DB ID>
is_a: <parent id> ! <parent name>
PRO term name
The Term name at this level follows UniProtKB naming guidelines as described in section 2.2.
PRO term definitions
Terms in this metaclass are typically defined with respect to a gene in a reference organism, and the class of proteins covered by the term includes all translation products not just of that gene in that organism, but also of any of its 1:1 orthologs in other organisms.
"A <parent term> that is a translation product of the <organism of reference> <gene name> gene or a 1:1 ortholog thereof."
For KCNA5 (PR:000000812):
"A voltage-gated potassium channel alpha subunit that is a translation product of the human KCNA5 gene or a 1:1 ortholog thereof." Additional information may be added.
The gene name from UniProtKB/Swiss-Prot is used since the gene name field is curated, providing standardization at least within certain taxon groups (vertebrates, plants, fungi, prokaryotes). PRO uses this source for gene names. We created an arbitrary priority on the gene name selected for the definition mammalian>prokaryote>fungi>plant>others. This priority is mainly based on the availability of a consistent source of gene names. Whenever there is a human protein product, we use the gene name in UniProtKB (which comes from HGNC) as a reference for the gene, and if other names are known these are added as synonyms. If there is a eukaryotic representative, gene names should be in upper case. However, if there is a prokaryotic only class, then we adopt the Demerec nomenclature.
Example, for cytochrome c (PIRSF500152):

Fig.3. Partial snapshot of members of the cytochrome C family (PIRSF500152) and the gene name of its members.
In this case the gene name CYCS is used as primary in the definition (CYCS) and the alternative ones are added as synonyms (CYTC, CYC1, CYC). Note: synonyms of genes should be added in the protein term as Related synonyms, since they are not protein names.\
If the group that is being under annotation has no mammalian representative, like homoserine dehydrogenase:

Partial snapshot of members of the homoserine dehydrogenase family (PIRSF000098), and the gene name of its members. This query has been performed in the PIR website
In the snapshot above the gene name "hom" is used as primary and others as synonyms (e.g. thrA). We include OrderLocusNames or ORFNames only in species specific terms.
There are cases of proteins that present in a specific organism for which there is no sequenced genome, gene name, etc, like latrotoxin (P23631, http://purl.obolibrary.org/obo/PR_000001088), in this case the definition is of the type:
"A protein that is a constituent neurotoxin of the black widow spider venom, and contains multiple ankyrin repeats (20-22)."
Each definition has a source that needs to be added (PMID, curator, Wikipedia, etc). In the case of the curator the format is PRO:<curator initials>.
e.g.
[Term]
id: PR:000000812
name: voltage-gated potassium channel subunit KCNA5
def: "A voltage-gated potassium channel alpha subunit that is a translation product of the human KCNA5 gene or a 1:1 ortholog thereof. The voltage-gated potassium channel Kv1.5 mediates the ultrarapid-activating potassium current (IKur) in heart, and also plays a critical role in the regulation of arterial tone." [PMID:10575199, PMID:16772329, PMID:18344374]
comment: Category=gene.
synonym: "HPCN1" EXACT []
synonym: "KCNA5" EXACT PRO-short-label [PRO:DNx]
synonym: "KV1-5" EXACT []
synonym: "voltage-gated potassium channel HK2" EXACT []
synonym: "voltage-gated potassium channel subunit Kv1.5" EXACT []
xref: PANTHER:PTHR11537\:SF25
is_a: PR:000000709 ! shaker-related voltage-gated potassium channel alpha subunit
Category=Organism-gene
This class is species-specific, the terms are children of the gene level.
[Term]
id: PR:<ID>
name: <parent name> (<organism>)
def: "A <parent> that is encoded in the genome of <organism>." [PRO:<curator initials>, PMID:<number>, <DBname>:<DB ID>]
comment: Category=gene.
synonym: "<name>" <EXACT/RELATED/BROAD/NARROW> []
xref: <DB name>:<DB ID>
intersection_of: <parent id> ! <parent name>
intersection_of: only_in_taxon NCBITaxon<id> ! <organism name>
relationship: has_gene_template DBID:<ID> ! name
PRO ID
The PRO ID conforms to PR:<UniProtAc> whenever there is a correspondence with UniProtKB.
PRO term name
In general the PRO term name conforms to the parent name followed by the organism name in parenthesis. However, there may be cases where the parent and children name may differ if the convention for naming the gene product is different for the given organism.
PRO term definition
The definition follows the format:
"A <parent> that is encoded in the genome of &llt;organism>"
e.g. http://purl.obolibrary.org/obo/PR_000025342
These terms are also defined logically as the intersection of the parent and the organism terms. If the organism is from a model organism database (MOD), then the gene template is also indicated with relationship has_gene_template and with link to the MOD.
[Term]
id: PR:P22460
name: potassium voltage-gated channel subfamily A member 5 (human)
def: "A voltage-gated potassium channel subunit KCNA5 that is encoded in the genome of human." []
comment: Category=organism-gene.
synonym: "hKCNA5" EXACT PRO-short-label [PRO:DNx]
xref: UniProtKB:P22460
intersection_of: PR:000000812 ! voltage-gated potassium channel subunit KCNA5
intersection_of: only_in_taxon NCBITaxon:9606 ! Homo sapiens
relationship: has_gene_template HGNC:6224
Cross-reference
Commonly Xref line sources in this category are from UniProtKB and/or Reactome.
xref: UniProtKB:P22460
xref: Reactome:REACT_25495
3.2-ProForm terms
The ProForm level encompasses the categories sequence and modification.
The ontology at the ProForm level reflects the conservation of specific isoforms and modified forms among species. We have introduced two terms to refer to these: 'ortho-isoforms' (orthologous isoforms) and "ortho-modified forms ."
Isoforms-encoded by orthologous genes-that are believed to have arisen prior to speciation and divergence of the primary sequence. That is, ortho-isoforms were true alternative isoforms (as defined above) in a common ancestor, and quite likely functionally equivalent.
Ortho-modified forms indicate the PTMs on ortho-isoforms that occur in equivalent residues. E.g. PR:000000650.
3.2.1 Curation of Category=sequence
Each PRO term at this level refers to the protein products with a distinct sequence upon initial translation. The sequence differences can arise from different alleles of a given gene, from splice variants of a given RNA, or from alternative initiation and ribosomal frameshifting during translation. One can think of this as a mature mRNA-level distinction. For example, smad2 encodes both a long splice form and a short splice form. The
protein products from each isoform are separate PRO terms. Sequence-level distinction is the first (parent-most) node of the ProForm part of PRO.
Isoforms and sequence variants are created based on information in manually curated databases such as UniProtKB entries and literature. Only variant related to disease are considered (see below). The curated ProForm terms are based on the experimentally observed entities
a-Isoforms
Isoforms are children of gene category terms. The source of isoform information comes from UniprotKB ( alternative product section) or the literature. For different organisms we create a single isoform term when we consider the protein to be orthologous isoforms. We are quite confident in considering equivalent isoforms those giving protein products with end to end similarity and no gaps. Also there may be cases where transcript with different UTR regions will give raise to the same protein sequence, these are all considered as one protein form.
Figure 5 shows the BLAST result (against mouse proteins) of two isoforms of the human TGF-beta receptor type-2. It shows the corresponding isoforms (P37173-1 equivalent to Q62312-2; P37173-2 equivalent to Q62312-1). The sequence matches with no gaps.
More in depth analysis is needed when there are small gaps in the sequence (1-3 aa long) to see if this is accounted for species diversity or a different forms. However, literature descriptions of an isoform in the different organisms are the best source for unifying terms.
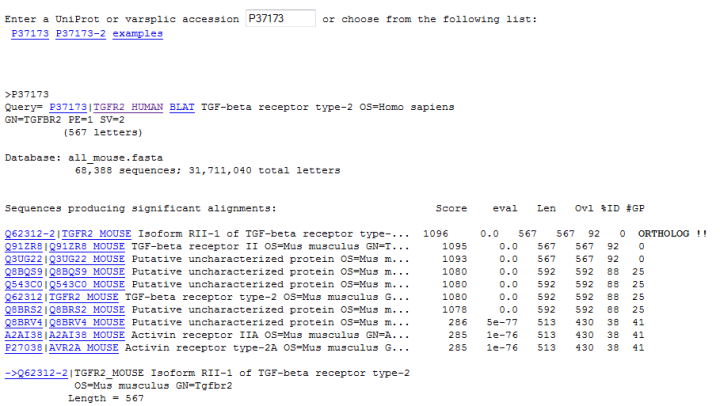

Fig.8. Snapshot of preliminary PRO terms for TRPC4 based on human and mouse protein entries.
In this case, all the isoforms extracted from the protein records are kept separate. However, further analysis of the corresponding isoform shows that these could be considered equivalent (sequence alignment, none of the existing mouse sequences containing the ESSN missing sequence, also when performing a BLAT of the human sequence against the mouse genome there is no match within this region. The alignment with rat below shows that this region is diverse among the different organisms. But most importantly, the literature backs up this information (PMID:11713258: "Like the murine beta splice variant, rat and human TRPC4beta both formed receptor-regulated cation channels when expressed in HEK293 cells. In contrast, human TRPC4alpha was poorly activated by stimulation of an H(1) histamine receptor").

Fig.9. Alignment of TRPC4 isoform beta in human, rat and mouse. The red box indicates the region where there is a gap in the sequence. The right box shows the alignment of this region with the mouse genome (using BLAT). The alignment shows that this is a continuous region in the sequence and does not constitute a different exon . The same is true if the mouse sequence is aligned against the human genome.
So in this particular case, the isoforms alpha and beta in mouse are equivalent to the corresponding isoforms in human and then only one ortho-isoform term should be created. Then a total of 4 isoforms alpha-delta are listed.
If there is no clear knowledge on the equivalency of isoforms, then they should not be created at the sequence level, but as organism-sequence level as a child of the organism-gene level.
Definitions of isoforms:
Definitions have to be consistent we use the format where we refer to a reference sequence, one that is the source of the experimental information.
Definition of isoforms follows the form:
A <parent name> that is a translation product of a mature transcript of the <gene name> gene. Represented by the sequence UniProtKB:<AC-#>, and its ortho-isoforms. Additional information can be added afterwards.
Where the < gene name> should be the same as the parent node, and the UniProtKB accession should be in the form of Q13616-1. Add -1 even when there is no other characterized isoform.
Whenever there is some information from literature about a specific sequence feature of an isoform, this should be included in the definition:
A TGF-beta receptor type-2 that is a translation product of a mature transcript of the TGFBR2 gene. Represented by the sequence UniProtKB:Q62312-1, and its orthoisoforms. This form has an insertion of approximately 25 amino acids in the ectodomain just adjacent to the signal peptide.
A sara that is a translation product of a mature transcript of the ZFYVE9 gene. Represented by the sequence UniProtKB:O95405-3, and its ortho-isoforms. Lacks the smad-binding domain.
A cullin 1 that is a translation product of a mature transcript of the CUL1 gene. Represented by the sequence UniProtKB:Q13616-1, and its ortho-isoforms.
We try to avoid mentioning exons since this is very species specific, though in some cases it could be used if the appropriate source for the nomenclature is added.
A rho-associated protein kinase 1 that is a translation product of a mature transcript of the ROCK2 gene, represented by the mouse sequence UniProtKB:…. Contains the core domains but with an inclusion of a sequence from between the rho binding domain and the PH domain (exon 27' in mouse).
b-Sequence variants
UniProtKB lists sequence variants mainly for human entries, there are few mouse entries with sequence variants which are attributed to different mouse strains. This doesn't prevent the creation of new terms based on mouse variants based on literature.
Only variants with a link to an OMIM phenotype record are considered in the ontology, so there may be cases where terms need to be deleted.
Special attention is needed to the vocabulary used in the CC disease line to see whether the sequence is "a cause of" or if it is "the cause of" a given disease. The former expression is used when the disease is/may be caused by mutations in different genes (locus heterogeneity) or mutations in 1 gene are not sufficient for disease manifestation, whereas the latter expression is used when the disease is/may be caused by mutations in one given gene only.
This will determine when to use the relationship agent_in (only in the last case).
Variants should be listed under the organism-gene specific term. An umbrella variant term is created
The standard definition used for the umbrella variant term is:
A <parent name> that has at least one amino acid residue that differs from the corresponding residue found in another <parent name> when such a difference arises as the result of genetic differences, i.e., different alleles of <parent term>. Operationally, this means the sequence representation of the <parent term> has at leas one amino acid residue that differs from the corresponding residue found in the sequence representation given by UniProtKB:<Ac-canonical sequence>. Add extra info if necessary.
E.g.
[Term]
id: PR:000025558
name: amyloid beta A4 protein sequence variant (human)
def: "An amyloid beta A4 protein (human) that has at least one amino acid residue that differs from the corresponding residue found in another amyloid beta A4 protein (human) when such a difference arises as the result of genetic differences, i.e., different alleles of amyloid beta A4 protein (human). Operationally, this means the sequence representation of the amyloid beta A4 protein (human) has at least one amino acid residue that differs from the corresponding residue found in the sequence representation given by UniProtKB:P05067-1." [PRO:DAN]
comment: Category=organism-sequence.
is_a: PR:000025532 ! amyloid beta A4 protein (human)
The children of this term are the specific variants and the definition follows:
A <parent term> that has <Aaa> residue at the position equivalent to <Aaa-#> in the <species name> sequence UniProtKB:<Acc-cannonical>.
An amyloid beta A4 protein sequence variant (human) that has an Asn residue at the position equivalent to Asp-678 in the human sequence UniProtKB:P05067-1." re generic definition when the variation takes place in a specific residue within a conserved feature. See:
3.2.2 Curation of Category=modification
Each PRO term at this level refers to the protein products derivedfrom a single mRNA species that differ because of some change (or lack thereof) that occurs after the initiation of translation (co- and post-translational). This includes sequence differences due to cleavage and chemical changes to one or more amino acid residues. For example, the long isoform of smad2 can either be unmodified or be post-translationally modified to contain phosphorylated residues. Modification-level distinction is the leaf-most node of the ProForm part of PRO. The modified forms are the proteins which have undergone cleavage or co/post translational modification (PTM). We do not intend to provide the time space information, i.e., the order on which modifications occurs, protein ontology only lists the final modified objects which are relevant for function.
is_a isoform 1
is_a isoform 1 unphosphorylated 1
is_a isoform 1 phosphorylated 1
is_a isoform 1 phosphorylated 2
is_a isoform 1 unphosphorylated 2
When various PTMs coexist, the modifications are listed in alphabetical order
is_a isoform 1
is_a isoform 1 acetylated, phosphorylated and ubiquitinated 1
There is a control vocabulary for the modifications (see document at the end of this file CREATE DOCUMENT)
We add the corresponding Evidence code in comment line if the forms have been observed in vitro (ECO:0000181) or in vivo (ECO:0000178).
[Term]
id: PR:000037408
name: transcription factor SPEECHLESS phosphorylated 1 (Arabidopsis thaliana)
def: "A transcription factor SPEECHLESS that has been phosphorylated in several Ser or Thr residues with then MAPK consensus motif Px[ST]P in Arabidopsis thaliana. Example: UniProtKB:Q700C7-1, Ser-193, MOD:00046|Ser-211, MOD:00046|Thr-214, MOD:00047|Ser-219, MOD:00046|Ser-255, MOD:00046." [PRO:LVM, PMID:19008449]
comment: Category=organism-modification. Evidence=(ECO:0000181).
is_a: PR:Q700C7 ! transcription factor SPEECHLESS (Arabidopsis thaliana)
relationship: only_in_taxon NCBITaxon:3702 ! Arabidopsis thaliana
As the starting point, the information about PTMs comes from the CC PTM and the FT lines in UniProtKB report. In addition, other useful resources are Phosphosite, Reactome, PUBMED and RLIMS-P (https://proconsortium.org/pirwww/iprolink/rlimsp.shtml).
Important considerations:
Make sure you are annotating to the correct isoform. Look for source of sequence, especially to constructs described in Materials and Methods, or mentioned in Acknowledgements, supplementary material, sequences shown in figures. Sometimes you need to trace back many papers to get to the source of the sequence (isoform, organism, etc. See Fig.10).
If you are still not sure which the isoform is, then you should contact the authors to request clarification.
To see if a specific form is identified, for example a specific phosphorylated form, check in the Material and methods section about the antibody used. There are certain specific antibodies raised against a given phosphorylated form, like the anti-phosphoSmad2 which recognizes the form that is phosphorylated at the two most C-terminal Ser residues (see example below).
It is important to note that the modified sites described in the protein forms are the ones known to have some functional implication.

!worddavdc7bb4fecc1c6d0f2687917147e6a0a0.png|height=51,width=700!PMID: 17074756 - The DNA binding activities of Smad2 and Smad3 are regulated by coactivator-mediated acetylation. PMID: 9738456
Fig.10. Two different papers have to be looked at to find the information for the antibody recognizing phospho-Smad2.
Keep in mind that the information in UniProtKB about PTMs is just a listing and does not provide the PTM combination that could be found in the protein form. In PRO we try to describe the observed form, so if this form is known to be acetylated and phosphorylated, as has been identified in the paper above for Smad2 isoform 2 (known as deltaexon3) we list it as one term.

Fig.11 Ontology for smad2 isoform 2.
Definitions for the umbrella terms follow the structure:
For unmodified forms:
A <parent name> that has not been subjected to <modification> at sites analogous to x, y, z,... n in UniprotKB:<accession>. Example: UniProtKB:<accession>, Aaa-#, PR:000026291.
Where PR:000026291 indicates unmodified residue.
For modified forms:
A < parent name> that has been post-translationally modified to include PTM-1, PTM-2,….PTM-X residues.
For cleavage
A <parent name> that has been processed by proteolytic cleavage.
Each specific modified form, child of the umbrella term, is created based on literature. Only the modifications that are relevant for function are described, at least for now. Add MOD, GO, SO term that is more appropriateThis section is not part of the ontology in the PAF file.
Even when the modified residues are not identified in the paper, a term can be created, as long as there is a way to define it.
For example, if there is a phosphorylated form that is inactive, that happens through the activation of a conserved pathway o kinase, then this can be used to define this form.
Example:
A smad2 isoform 1 phosphorylated form that has been phosphorylated at [S/T] residues within the MH1-MH2 domain linker region in response to decorin-induced Ca(2+) signaling.
Gene fusion products
A protein that is a translation product of the fusion of (HGNC gene 1 symbol) and (HGNC gene 2 symbol) genes.
3.3 ProComp Curation
ProComp represents protein-containing complexes including protein-only and protein-other macromolecule complexes. The root is GO protein-containing complex (GO:0032991) defined as “A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.”
Protein complexes are distinguished from protein-protein interactions in that they are continuant entities, i.e. they endure or continue to exist through time. Interactions, in contrast, are occurrent entities, i.e. they occur in time through successive temporal phases (PMID:21929785).
Note that a protein in monomeric state linked non-covalently to a small chemical is not part of ProComp, but it can be defined as a subset of the protein term with has_bound CHEBI ID. See example below for GDP bound form of Ran. In this cases we still use as category the complex level.
[Term]
id: PR:#####
name: GTP-binding nuclear protein Ran, GDP-bound form (human)
def: "A GTP-binding nuclear protein Ran (human) that is non-covalently bound to GDP." [PDB:3GJ0]
comment: Category=organism-complex .
synonym: "human RanGDP" EXACT [PDB:3GJ0]
intersection_of: PR:P62826 ! GTP-binding nuclear protein Ran (human)
intersection_of: bound_to CHEBI:17552 ! GDP
PRO represents complexes explicitly, defining each member of the complex at the level of its isoform, variant, or modified form, whenever possible. This provides the ability to represent complex biological knowledge as it is emerging in the experimental research community in structures that are both human readable and accessible to algorithmic approaches.
ProComp leverages, and cross references, entries in existing protein-centric informatics resources, including the protein-containing complexes that are represented in the Cellular Component branch of the Gene Ontology. In the GO, types of protein-containing complexes are defined in terms of constituent molecule classes and the function(s) that the complexes carry out. By agreement within the protein informatics community, PRO represents the species-specific classes of protein complexes, while GO, in most instances, represents the species-independent classes of protein complexes; within PRO, the latter are referred to by using GO identifiers.
ProComp stanza
The protein complex types in PRO are defined by means of multiple has_component relationships to PRO-defined species-specific protein types. The species-specific protein types can be isoforms or modified isoforms. Each PRO complex stanza includes an is_a relationship to the corresponding Gene Ontology (GO) protein complex term or a PRO term when no GO term can be made. The species specificity of the protein complex is indicated by the value of the only_in_taxon tag. The has_component relationships assert that each instance of the complex contains an instance of indicated peptide or compound. The embedded cardinality declaration indicates the number of instances of that type of subunit in the complex. The absence of a cardinality declaration indicates there is at least one copy of each subunit.
Example for organism-specific
[Term]
id: <PR ID>
name: <term name> (organism name)
def: <definition> [<evidence source>]
comment: Category=organism-complex.
xref: <DB if applies>
is_a: <GO complex ID/PR complex ID>
relationship: has_component <PR ID1> {cardinality="<number, optional>"} ! <PR term name 1>
relationship: has_component <PR IDn> {cardinality="<number, optional>"} ! <PR term name n>
relationship: only_in_taxon NCBITaxon:<taxID> ! <organism name>
Real Example:
[Term]
id: PR:000026434
name: DNA replication factor C complex (human)
def: "A DNA replication factor C complex whose components are encoded in the genome of human." [PRO:DAN, Reactome:REACT_4881]
comment: Category=organism-complex.
synonym: "RFC heteropentamer (human)" EXACT [Reactome:REACT_4881]
is_a: GO:0005663 ! DNA replication factor C complex
relationship: has_component PR:P35250 {cardinality="1"} ! replication factor C subunit 2 (human)
relationship: has_component PR:P40938 {cardinality="1"} ! replication factor C subunit 3 (human)
relationship: has_component PR:P35249 {cardinality="1"} ! replication factor C subunit 4 (human)
relationship: has_component PR:P35251 {cardinality="1"} ! replication factor C subunit 1 (human)
relationship: has_component PR:P40937 {cardinality="1"} ! replication factor C subunit 5 (human)
relationship: only_in_taxon NCBITaxon:9606 ! Homo sapiens
Species non-specific terms
All complexes in ProComp should be given parentage under the general term protein-containing complex; GO:0032991. PRO species non-specific terms should be GO terms whenever possible. There are some cases where GO terms won’t be created for species non-specific complexes so we create the corresponding PRO terms. We should request GO complex terms as needed via the GO issue tracker (https://github.com/geneontology/go-ontology/issues/), providing some background information (PMIDs) and, especially, showing that the complex is conserved in some taxonomic range.
Example of a PR term for species non-specific term.
GO:0046696 “A multiprotein complex that consists of at least three proteins, CD14, TLR4, and MD-2, each of which is glycosylated”
Note that there are many “flavors of CD14” soluble vs. membrane-bound and also whether LPS is bound to MD-2 or CD14 in the complex. So we have in PRO 3 children complexes of this term which are species non-specific. In these cases we add the has_component to the stanza.
Example:
[Term]
id: PR:000025497
name: lipopolysaccharide receptor complex 3
def: "A lipopolysaccharide receptor complex that contains the membrane bound form of CD14, and the lipopolysaccharide (LPS) bound to MD-2." [PMID:7539760, PMID:9464818]
comment: Category=complex.
is_a: GO:0046696 ! lipopolysaccharide receptor complex
relationship: has_component CHEBI:16412 ! lipopolysaccharide
relationship: has_component PR:000002149 ! CD14 molecule isoform 1, signal peptide removed cleaved and GPI-anchored 1
relationship: has_component PR:000003299 ! lymphocyte antigen 96 isoform 1, signal peptide removed glycosylated 1
relationship: has_component PR:000025492 ! toll-like receptor 4 isoform 1, signal peptide removed glycosylated 1
Note that the change in hierarchy in GO for lipopolysaccharide receptor complex term has been requested (Ticket 10255, change parenthood to macromolecular complex).
Species-specific terms
As mentioned in previous sections species-specific terms have in most cases GO terms as parent.
Example:
[Term]
id: PR:000028439
name: RAD51:BRCA2 complex (human)
def: "A protein complex whose components are one molecule of breast cancer type 2 susceptibility protein (human), and one molecule of DNA repair protein RAD51 1 (human)." [PRO:PDE, Reactome:REACT_4224]
comment: Category=organism-complex.
xref: Reactome:REACT_4224
is_a: GO:0043234 ! protein complex
relationship: has_component PR:000027310 {cardinality="1"} ! DNA repair protein RAD51 1 (human)
relationship: has_component PR:000028399 {cardinality="1"} ! breast cancer type 2 susceptibility protein (human)
relationship: only_in_taxon NCBITaxon:9606 ! Homo sapiens
In PRO it is possible to define a complex based on its subcellular location when this confers some functional difference. These complexes are made as subset of the standard complex term and the definition includes intersection to GO subcellular component term. In this case we indicate the localization in the PR name separated by semicolon
<PR name>; <subcell location> (<organism>)
Example:
The LPS receptor complex 3 participates only in the MyD88-independent toll-like receptor signaling pathway when located in the endosomal membrane (PR:000036137), whereas in plasma membrane it has the potential to participate in the MYD88-dependent or independent pathways.
Here is an example with the formal definition
[Term]
id: PR:000036137
name: lipopolysaccharide receptor complex 3; endosome membrane (human)
def: "A lipopolysaccharide receptor complex 3 (human) that is localized to the endosome membrane." [Reactome:REACT_124771]
comment: Category=organism-complex.
synonym: "TLR4:MD2:LPS:CD14 [endosome membrane] (Homo sapiens)" EXACT [Reactome:REACT_124771]
xref: Reactome:REACT_124771
intersection_of: PR:000025773 ! lipopolysaccharide receptor complex 3 (human)
intersection_of: located_in GO:0010008 ! endosome membrane
Evidence codes
Evidence codes are added in comment line with format:
Evidence=(<ECO ID>, based on <cite source>).
Use evidence code when:
-Protein Complexes are inferred by homology
We can create PRO terms for inferred complexes only when all the components are conserved. Although be mindful of data regarding the protein component localization/expression. If the inferred complex components are In this case we add the Evidence code ECO:0000088 (biological system reconstruction) in the comment line with the following format:
Evidence=(ECO:0000088, based on <PR ID of experimentally determined homolog complex>).
[Term]
id: PR:000027758
name: 4-aminobutyrate aminotransferase (mouse)
def: "A 4-aminobutyrate aminotransferase whose components are encoded in the genome of mouse." [PRO:CJB]
comment: Category=organism-complex. Evidence=(ECO:0000088, based on PR:000027757).
synonym: "GABA-T (mouse)" EXACT [MGI:2443582]
is_a: GO:0032144 ! 4-aminobutyrate transaminase complex
relationship: has_component PR:000027700 {cardinality="2"}! 4-aminobutyrate aminotransferase, mitochondrial isoform 1 (mouse)
relationship: only_in_taxon NCBITaxon:10090 ! Mus musculus
-Protein Complexes are inferred by system reconstruction
Sometimes these data are generated in in vitro reconstitution systems that may include proteins derived from multiple sources and perhaps different species. Although we will try to avoid to generate PRO terms for this type of complexes, if you do please add
Evidence=(ECO:0000088, based on <cite source>).
We will try to avoid curating these complexes, and do this mainly for requests and when needed for human/mouse comparison.
-Protein Complexes are asserted by curator
We will use ECO:0000305 (curator inference used in manual assertion ) for cases where we make assertions of complexes and/or their components based on additional knowledge than that explicitly provided in the references. E.g., in some complexes we know that the activated form of the complex includes some particular phosphorylated form of a protein, though the paper may not show that explicitly.
Format:
Evidence=(ECO:0000305, based on <cite evidence>).
3.4-Format of comment line in PRO terms
The comment line is free text but in PRO comments are semi-structured by type of comments. Each comment has a heading and is separated by period.
Category:
This field is meant to indicate the level of granularity in the ontology.
The first comment is Category. All PRO terms, except those obsoleted, have a Category comment with format:
Category=<name>.
It is controlled vocabulary for this. Below is the list of possible categories
- Category=family.
- Category=gene.
- Category=organism-gene.
- Category= sequence.
- Category=organism-sequence.
- Category= modification.
- Category=organism-modification.
- Category=union.
Evidence:
This field is meant to indicate the evidence for term created. We use ECO: http://bioportal.bioontology.org/ontologies/ECO?p=classes&conceptid=root
Evidence=(<ECOID>, based on <ID>).
Evidence generally used:
ECO:0000088 biological system reconstruction. Evidence based on reconstruction of a biological system that is based on an existing experimentally supported model of that biological system.
ECO:0000250 sequence similarity evidence used in manual assertion.
ECO:0000205 curator inference. An evidence type that is based on conclusions drawn by a curator.
ECO:0000006 experimental evidence (EXP). An evidence type that is based on the results of a laboratory assay.
ECO:0000311 An evidence type that is based on work performed by a person or group prior to a use by different person or group.
ECO:0000322 imported manually asserted information used in automatic assertion.
ECO:0000178 in vivo assay evidence
ECO:0000181 in vitro assay evidence
Evidence=(ECO:0000181, for kinase information). Evidence=(ECO:0000178, for phosphorylation).
http://purl.obolibrary.org/obo/PR_000044375
Notes:
This field is meant to provide clarification or comments about the term.
Note: <text>.
Space then start in lower case.
http://purl.obolibrary.org/obo/PR_000037803
Modifying enzymes:
This field is meant to indicate the PTM enzyme(s) if known. Kinase, Acetylase, etc
Kinase=("<name>"; <PROID>; "site1-#", "site2-#).
For name we normally use the gene name or complex name.
Kinase=("DAPK1"; PR:P53355; Ser-309, Ser-311, Ser-312, Ser-318, Ser-326).
http://purl.obolibrary.org/obo/PR_000037784
Requested by
This field is meant to indicate the institution that requested the term.
Requested by=TLR.
4-References
Day-Richter J., Harris M.A., Haendel M.; Gene Ontology OBO-Edit Working Group, Lewis S. (2007) OBO-Edit--an ontology editor for biologists.Bioinformatics, 23:2198-2200.
Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, Kitano H, Thomas PD (2005) The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res., 33:D284-288.
Wu C.H., Nikolskaya A., Huang H., Yeh L-S., Natale D.A., et al. (2004) PIRSF family classification system at the Protein Information Resource. Nucleic Acids Res., 32:D112-114.
5-Resources
Data sources:
GO: http://www.geneontology.org/
MGI: http://www.informatics.jax.org/
MIM: http://www.ncbi.nlm.nih.gov/sites/entrez?db=omim
Pfam: http://pfam.sanger.ac.uk/
PIRSF: https://proconsortium.org/pirwww/dbinfo/pirsf.shtml
PSI-MOD: http://psidev.sourceforge.net/mod/
SO: http://www.sequenceontology.org/
UniProt: http://beta.uniprot.org
Useful Resources:
HPRD: http://www.hprd.org/index_html
Phosphosite:http://www.phosphosite.org
NCBI resources: http://www.ncbi.nlm.nih.gov/
UCSC Genome Bioinformatics (genome browser, BLAT): http://genome.brc.mcw.edu/

